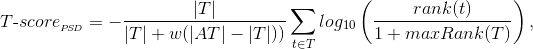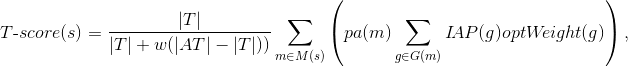| ID |
Name |
Target names |
Target activity score |
NA |
Phase 1 |
Phase 2 |
Phase 3 |
Phase 4 |
Disease activity score |
Drug rank |
| Regorafenib |
KIT, KDR, ABL1, PDGFRB, FGFR1, RET, PDGFRA... |
2 |
Colorectal Neoplasms, Adenocarcinoma, Carcinoma, Hepatocellular, Cholangiocarcinoma, Gastrointestinal Stromal Tumors, Glioblastoma, Liver Neoplasms... |
Colorectal Neoplasms, Carcinoma, Hepatocellular, Carcinoma, Small Cell, Esophageal Neoplasms, Gastrointestinal Neoplasms, Gastrointestinal Stromal Tumors, Intestinal Neoplasms... |
Colorectal Neoplasms, Adenocarcinoma, Bile Duct Neoplasms, Brain Abscess, Breast Neoplasms, Carcinoid Tumor, Carcinoma, Adenoid Cystic... |
Colorectal Neoplasms, Carcinoma, Hepatocellular, Colonic Neoplasms, Esophageal Neoplasms, Gastrointestinal Stromal Tumors, Neoplasms, Noma... |
Colorectal Neoplasms, Gastrointestinal Stromal Tumors, Neoplasms, Rectal Neoplasms |
11 |
4 |
| Sorafenib |
KIT, KDR, PDGFRB, FGFR1, BRAF, RAF1, RET |
1.49 |
Colorectal Neoplasms, Adenocarcinoma, Ascites, Brain Abscess, Brain Neoplasms, Breast Neoplasms, Carcinoma, Hepatocellular... |
Colorectal Neoplasms, Adenocarcinoma, Adenoma, Adenoma, Liver Cell, Astrocytoma, Bile Duct Neoplasms, Biliary Tract Neoplasms... |
Colorectal Neoplasms, Adenocarcinoma, Adenoma, Adenoma, Liver Cell, Adrenocortical Carcinoma, Bile Duct Neoplasms, Biliary Tract Neoplasms... |
Adenocarcinoma, Breast Neoplasms, Carcinoma, Carcinoma, Hepatocellular, Carcinoma, Non-Small-Cell Lung, Carcinoma, Renal Cell, Digestive System Diseases... |
Carcinoma, Hepatocellular, Carcinoma, Renal Cell, Liver Neoplasms, Neoplasms, Noma, Thrombosis |
4 |
25 |
| Nintedanib |
FGFR3, SRC, KDR, LYN, FGFR1 |
1.08 |
Colorectal Neoplasms, Carcinoma, Carcinoma, Non-Small-Cell Lung, Endometrial Neoplasms, Fallopian Tube Neoplasms, Idiopathic Pulmonary Fibrosis, Lung Diseases... |
Adenocarcinoma, Breast Neoplasms, Carcinoma, Hepatocellular, Carcinoma, Non-Small-Cell Lung, Carcinoma, Renal Cell, Carcinoma, Small Cell, Colonic Neoplasms... |
Colorectal Neoplasms, Adenocarcinoma, Adenocarcinoma, Clear Cell, Adenocarcinoma, Mucinous, Angiomyoma, Appendiceal Neoplasms, Breast Neoplasms... |
Colorectal Neoplasms, Carcinoma, Non-Small-Cell Lung, Idiopathic Pulmonary Fibrosis, Lung Diseases, Lung Diseases, Interstitial, Mesothelioma, Neoplasms... |
Idiopathic Pulmonary Fibrosis, Pulmonary Fibrosis |
6 |
29 |
| Bosutinib |
CAMK2G, SRC, ABL1, MAP2K1, LYN |
1.55 |
Breast Neoplasms, Leukemia, Leukemia, Lymphoid, Leukemia, Myelogenous, Chronic, BCR-ABL Positive, Leukemia, Myeloid, Neoplasms, Precursor Cell Lymphoblastic Leukemia-Lymphoma |
Colorectal Neoplasms, Acute Kidney Injury, Breast Neoplasms, Carcinoma, Non-Small-Cell Lung, Cholangiocarcinoma, Cognitive Dysfunction, Dementia... |
Colorectal Neoplasms, Brain Abscess, Breast Neoplasms, Cholangiocarcinoma, Cysts, Glioblastoma, Kidney Diseases, Cystic... |
Leukemia, Leukemia, Myelogenous, Chronic, BCR-ABL Positive, Leukemia, Myeloid |
Leukemia, Myeloid |
3 |
31 |
| Dasatinib |
KIT, SRC, ABL1, PDGFRB, YES1, FYN, ABL2 |
1.29 |
Brain Neoplasms, Carcinoma, Squamous Cell, Carcinoma, Transitional Cell, Gastrointestinal Stromal Tumors, Glioblastoma, Leukemia, Leukemia, Lymphoid... |
Colorectal Neoplasms, Adenocarcinoma, Adenocarcinoma, Clear Cell, Adenocarcinoma, Mucinous, Brain Abscess, Brain Diseases, Breast Neoplasms... |
Colorectal Neoplasms, Adenocarcinoma, Adenocarcinoma, Clear Cell, Blast Crisis, Brain Abscess, Brain Diseases, Brain Neoplasms... |
Leukemia, Leukemia, Lymphoid, Leukemia, Myelogenous, Chronic, BCR-ABL Positive, Leukemia, Myeloid, Leukemia, Myeloid, Accelerated Phase, Leukemia, Myeloid, Acute, Leukemia, Myeloid, Chronic-Phase... |
Leukemia, Leukemia, Lymphoid, Leukemia, Myelogenous, Chronic, BCR-ABL Positive, Leukemia, Myeloid, Precursor Cell Lymphoblastic Leukemia-Lymphoma |
3 |
36 |